Showing posts with label compbio. Show all posts
Showing posts with label compbio. Show all posts
Thursday, April 29, 2010
ATP Metabolism
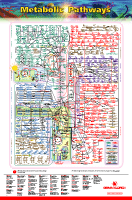
And on the subject of overwhelming biological data, this is the IUBMB-Nicholson chart of all the metabolic pathways that go into ATP management in mitochondria and chloroplasts, ATP being the basic energy currency of biological systems. There's a browser-crashing full sized pdf at the link, or click the above thumbnail for a jpg.
Labels:
biology,
compbio,
knowledge management
Friday, November 20, 2009
High Speed Sequencing
This video dedicated to my undergraduate degree in biology, in which it was never deemed necessary to introduce the fact that sequencing technology more sophisticated than the Sanger method exists. This is an animation explaining the process behind Helicos's new single-molecule sequencing technology. Like all other modern sequencing methods, this technique is based on short-read sequences-- DNA is replicated and then broken into millions of tiny fragments (25-50 base pairs at the low end), all of which are sequenced simultaneously. Given about 30-fold coverage of your genome, you can align these fragments to confidently reconstruct it as a single sequence.
Also of note, the Velvet algorithm is one cool sequence assembly program which, instead of aligning DNA fragments by simply looking for overlapping regions between them, plots all the fragment sequences generated onto a De Bruijn graph, and then uses principles of graph theory to condense them into a single sequence. Yay math!
Labels:
biology,
compbio,
computing,
pattern recognition,
technology
Sunday, October 11, 2009
Belousov-Zhabotinsky Reaction
And speaking of oscillations, here's a nice video of the Belousov-Zhabotinsky reaction in a petri dish:
(The video info for the above also has a nice description of waveforms in the brain which is worth reading.) You can see someone setting up the reaction (ie pouring some chemicals together and stirring, really) here.
The BZ reaction is an example of a chemical oscillator, a system which instead of arriving at a steady state transitions between two different chemical states (which two states have two different colors, whence the waves above). Boris Belousov discovered it in the 1950's when he happened to mix together potassium bromate, cerium(IV) sulfate, propanedioic acid and citric acid in dilute sulfuric acid (hell, why not?); he made two attempts to publish his findings, but was rejected from peer-reviewed journals because he couldn't explain why the oscillations occurred.
(The video info for the above also has a nice description of waveforms in the brain which is worth reading.) You can see someone setting up the reaction (ie pouring some chemicals together and stirring, really) here.
The BZ reaction is an example of a chemical oscillator, a system which instead of arriving at a steady state transitions between two different chemical states (which two states have two different colors, whence the waves above). Boris Belousov discovered it in the 1950's when he happened to mix together potassium bromate, cerium(IV) sulfate, propanedioic acid and citric acid in dilute sulfuric acid (hell, why not?); he made two attempts to publish his findings, but was rejected from peer-reviewed journals because he couldn't explain why the oscillations occurred.
Steven Strogatz's Sync
Steven Strogatz's wonderful book Sync discusses how synchrony in biological networks is not only common, but neigh-inevitable. His opening discussion of fireflies is particularly vivid: along some riverbanks in Southeast Asia, populations of fireflies stretching for miles will all flash in synchrony, a phenomenon which baffled western explorers for decades. It turns out the effect is easy to replicate in a model-- say you have a collection of periodic oscillators which fire a burst of light at their peak and then reset, you can achieve synchrony if you make it so that each oscillator, when it fires, bumps its neighbors forward a bit in their cycles. Because firing induces a forced reset of the cycle, oscillators will be pushed forward in their cycles until they fall into sync, and then stay locked there; this effect takes off in small groups and quickly grows until the entire network is firing together.
The important points here being that a) neurons do this too (in fact it can be hard to get spiking neural networks to stop doing this) and it's really probably important to coding somehow, and b) you guys, fireflies are totally attempting to form some sort of massive insect-based consciousness here.
You can read more on the subject in the preview of the first chapter and a half, posted on Google Books.
The important points here being that a) neurons do this too (in fact it can be hard to get spiking neural networks to stop doing this) and it's really probably important to coding somehow, and b) you guys, fireflies are totally attempting to form some sort of massive insect-based consciousness here.
You can read more on the subject in the preview of the first chapter and a half, posted on Google Books.
Monday, September 21, 2009
Computational Neuroscience Links
Computational Neuroscience on the World Wide Web-- a pretty comprehensive resource on comp neuro labs, tools, and conferences.
Labels:
compbio,
neuroscience
Friday, July 3, 2009
Protein Vaults
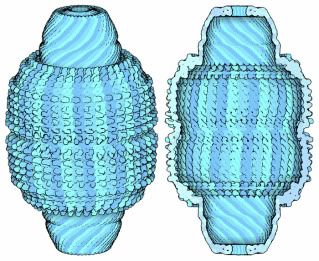
Vaults are the Protein Data Bank's molecule of the month. Nerdy, yes, but these sound kind of cool:
Vaults are composed of many copies of the major vault protein, which assembles to form a hollow football-shaped shell. The one shown here is from rat liver cells (PDB entries 2zuo, 2zv4, and 2zv5) and contains 78 copies of the protein. Inside cells, the vault also encloses a few other molecules, which were not seen in the crystal structure because they don't have a symmetrical structure inside the vault. These molecules include several small RNA molecules, a protein that binds to RNA, and an enzyme that adds nucleotides to proteins.
Labels:
compbio
23 Questions
Darpa's 23 mathematical challenges in modern scientific efforts, with a distinct emphasis on computational biology.
Larry Abbott's 23 questions in computational neuroscience.
23 is a reference to the father of all such lists, Hilbert's 23 problems posed to the International Congress of Mathematicians in 1900; these problems shaped much of 20th century mathematics, and some remain unresolved to this day.
Larry Abbott's 23 questions in computational neuroscience.
23 is a reference to the father of all such lists, Hilbert's 23 problems posed to the International Congress of Mathematicians in 1900; these problems shaped much of 20th century mathematics, and some remain unresolved to this day.
Labels:
compbio,
computing,
mad science,
math,
neuroscience,
research
Wednesday, May 6, 2009
Visualizing Music and Looking for Patterns
Found this and many other impressive videos on one Stephen Malinowski's YouTube channel. I really like the way the colored bar visualization separates out the different voices in a piece, especially the fugues.
The opening to Gödel, Escher, Bach has a fun discussion on the structure of the fugue-- the gist of it is that the composer develops the piece out of one short theme (a few measures of some simple melody), carried by a fixed number of voices. Starting with one voice expressing the theme, each additional voice chimes in repeating the theme until all are present. The theme is further explored and varied throughout the piece via transformations of the original melody: inverting, reversing, transposing, compressing. Soooo the video above is really cool, because the visualizations make it that much easier to pick out all the transformations that are taking place. Yay!
I wonder if you could make other visualization methods which help you pick out recurring themes in a piece, and are robust to transformations from the original theme (or measure distance from the original). It seems like a problem that crops up a lot, in problems from network analysis to predicting structural motifs in proteins. For instance, all integral membrane proteins will have a hydrophobic region which crosses the lipid bilayer-- this requires an extended sequence of hydrophobic amino acids, which will be reflected in the genetic code. There would be variation in sequence (not all membrane proteins would have the same arrangement of hydrophobic amino acids), but there might still be trends which might be picked up. Fourier/Laplace transforms can break a signal down into its periodic components; is there some way to transform a signal to visualize it in the space of its recurrent themes and their variations?
Labels:
compbio,
computing,
music,
pattern recognition,
videos
Sunday, October 19, 2008
Synthetic/Comp Bio links
Links to computational and synthetic biology-type blogs and such, from a family friend who follows such things. Some of these are a bit old or don't update much; The Seven Stones (the Molecular Systems Biology blog at nature.com) is the only one I've been any good at following consistently.
The Seven Stones
Synthesis
Paras Chopra
SyntheticBiology.org
Computational Systems Biology
Blogging the Biotech Revolution
nodalpoint.org
SynBERC
Beta Science
Suicyte Notes
The Loom
Homunculus
The Seven Stones
Synthesis
Paras Chopra
SyntheticBiology.org
Computational Systems Biology
Blogging the Biotech Revolution
nodalpoint.org
SynBERC
Beta Science
Suicyte Notes
The Loom
Homunculus
Subscribe to:
Posts (Atom)